Gibson Assembly Primers Design Tool
Primer Design and Fragment Assembly Using NEBuilder HiFi DNA Assembly® or Gibson Assembly®
Watch an interactive tutorial on primer design to see how simple it really is to clone with either NEBuilder HiFi DNA Assembly or the Gibson Assembly Cloning Kit.
Script
Building upon our introduction to NEBuilder HiFi DNA Assembly and Gibson Assembly, which detailed the versatility and power of these master mixes, we will now walk through the protocol for preparing fragments for assembly using either NEBuilder HiFi DNA Assembly or Gibson Assembly.
It is advisable to begin by assembling the final sequence in silico. This enables visualization of fragment junctions and facilitates the choice and design of the overlapping primers.
Design the nucleotide overhangs, which will enable correct fragment annealing. For optimal assembly, we recommend using fifteen- to forty-basepair overlaps that exhibit a melting temperature greater than 48 degrees Celsius.
For help with designing primers, use the NEBuilder Assembly Tool at nebuilder.neb.com. With NEBuilder HiFi DNA Assembly, if you increase the overlap region between fragments, you will increase efficiency and can use less DNA.
The basic steps of fragment assembly are as follows: First, design primers for your fragments. One of your primers will be designed to include a 15-40 base pair overlap with the primer sequence on the complementary strand.
Additional primer design approaches include adding the overlap region to the forward primer of Fragment B or splitting the overlap region between the reverse primer of Fragment A and the forward primer of Fragment B. After PCR, the resultant fragment includes the overlap region, and is now ready to be joined during Assembly.
These same basic steps can be used to clone an insert such a lacZ, into an expression vector, like pET21a, using PCR.
You will generate four primers: two forward primers and two reverse primers. To design the overlap of sequences, terminal regions of linearized pET21a sequence are added to the 5´ ends of the lacZprimers. Note that the forward primers share a region of complementarity with the reverse primers.
Following PCR with primers F2 and R2, the resultant lacZ gene sequence includes 15 to 40 nucleotide overlap regions with complementarity to the pET21a vector.
Finally, the fragments are joined by incubation with the Assembly Master Mix for 15 to 60 minutes at 50 degrees Celsius.
Following restriction enzyme digestion, vectors may have 5´ overhangs, 3´ overhangs, or blunt ends. The overlap region should always be generated by counting from the first nucleotide at the 3´ end, regardless of the type of overhang.
In this example, the lacZ gene is cloned into the pMAL-c5X vector, after digestion with NcoI and SbfI. The overlap region of the forward primer for the gene of interest should line up with the 3´ end of the overhang and extend back until the melting temperature of the overlap is greater than 48 degrees Celsius.
To ensure preservation of the reading frame, nucleotides may be added between the overlap region and the gene-specific sequence. A similar approach is then taken with the reverse primers.
After viewing these examples, you should now have an understanding of how to design primers to enable fragment assembly with either NEBuilder HiFi DNA Assembly or Gibson Assembly Master Mix.
Related Videos
-
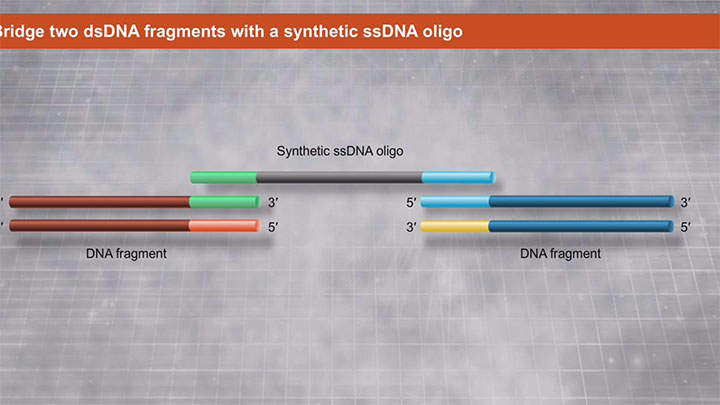
NEBUILDER® HIFI DNA ASSEMBLY ®: Bridging dsDNA with a ssDNA Oligo -

NEBUILDER HIFI DNA ASSEMBLY ® Removal of 3´ end Mismatches -
Introduction to NEBUILDER HIFI DNA ASSEMBLY ®
Visit NEB's Video Library
Gibson Assembly Primers Design Tool
Source: https://www.nebiolabs.com.au/tools-and-resources/video-library/primer-design-and-fragment-assembly-using-gibson-assembly
Posted by: elliottlizintacer1944.blogspot.com

0 Response to "Gibson Assembly Primers Design Tool"
Post a Comment